Transfer Learning Made Easy with Determined
September 01, 2023
In this blog post, we’ll explore how to use Determined to train a model on MedMNIST v2 – a widely-used lightweight benchmark for biomedical image classification – with 4 times the functionality and using half as many lines of code! To try this example out for yourself, check the GitHub examples repo and get in touch with us on the Determined Community Slack.
Deep learning based biomedical image analysis plays an increasingly important role in the healthcare industry. Recently, researchers published MedMNIST v2: A large-scale, lightweight benchmark for 2D and 3D biomedical image classification. This dataset contains several 2D and 3D datasets to train models to learn to classify colon pathologies, skin lesions, thorax diseases, and more. Since biomedical image analysis is an interdisciplinary field, it’s hard to find a dataset that is diverse yet standardized:
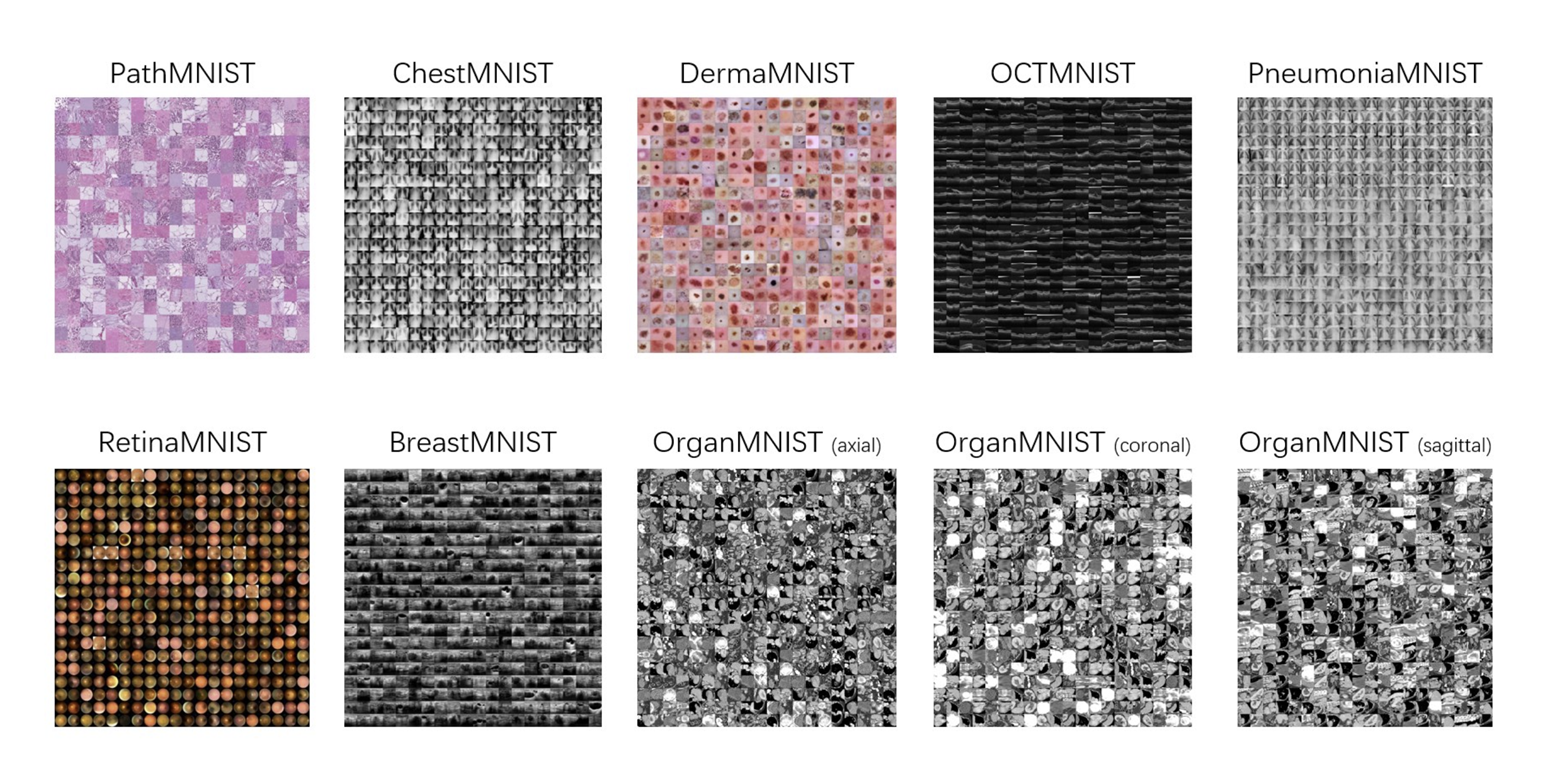
These datasets are perfect for transfer learning, or using a model trained on another dataset (like a ResNet pretrained on ImageNet) to learn another task. MedMNIST also contains sample training scripts to help you get started with these datasets, written using PyTorch, Keras, and SciKit-Learn. These training scripts illustrate the problem that biomedical researchers commonly face: it’s a pain to implement things that help scale up experimentation and make it more agile, like distributed training, hyperparameter search, checkpointing, visualization, and collaboration. Let’s take a look at how this training script handles some of the aforementioned components and how Determined makes it easier:
1) Distributed Training
In line 291, the gpu_ids are received from input arguments from the user:
gpu_ids = args.gpu_ids
The device is configured using only the first gpu_id (distributed training is not yet enabled):
device = torch.device('cuda:{}'.format(gpu_ids[0])) if gpu_ids else torch.device('cpu')
And the model is ported to the GPU in line 90:
model = model.to(device)
As well as the inputs to the model, for example, in line 190:
outputs = model(inputs.to(device))
The variable “device” is referenced a total of 27 times in the training script. This is a perfect example of how a researcher would normally need to manage training on a GPU – manually. And we haven’t even started distributed training yet!
With Determined, configuring distributed training is very simple:
resources:
slots_per_trial: 4
2) Visualization:
The original script uses a library called tensorboardX:
from tensorboardX import SummaryWriter
Using this library, a writer object is created for handling visualization data:
writer = SummaryWriter(log_dir=os.path.join(output_root, 'Tensorboard_Results'))
The writer object is referenced a total of 9 times throughout the script.
In addition, training and testing metrics are manually calculated and logged in various places throughout the script, e.g.:
logs = ['loss', 'auc', 'acc']
train_logs = ['train_'+log for log in logs]
val_logs = ['val_'+log for log in logs]
test_logs = ['test_'+log for log in logs]
log_dict = OrderedDict.fromkeys(train_logs+val_logs+test_logs, 0)
train_log = 'train auc: %.5f acc: %.5f\n' % (train_metrics[1], train_metrics[2])
val_log = 'val auc: %.5f acc: %.5f\n' % (val_metrics[1], val_metrics[2])
test_log = 'test auc: %.5f acc: %.5f\n' % (test_metrics[1], test_metrics[2])
log = '%s\n' % (data_flag) + train_log + val_log + test_log
print(log)
With Determined, all visualization is automatic:
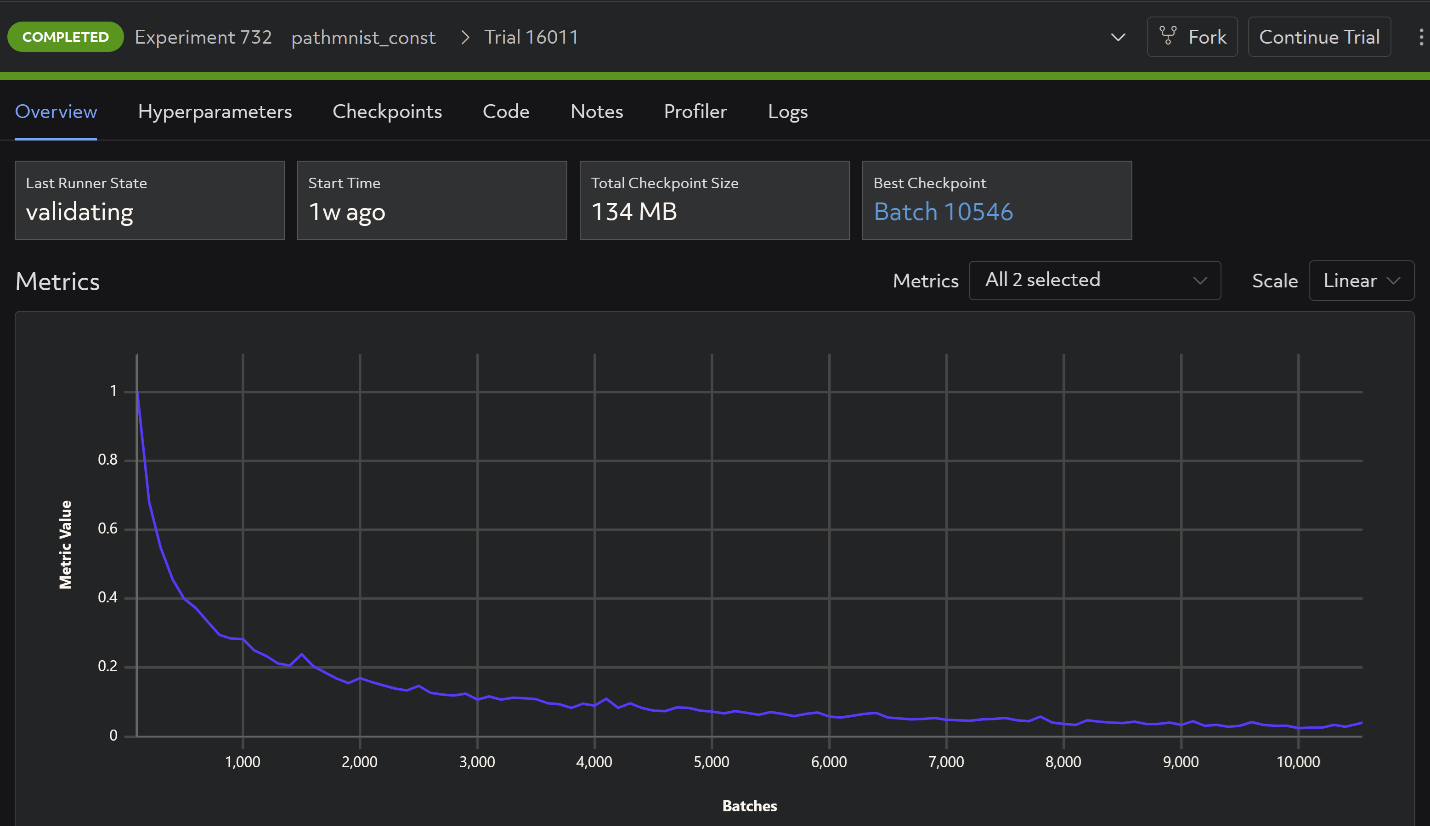
3) Checkpointing
The original training script saves the last model at the very end of the training process:
path = os.path.join(output_root, 'best_model.pth')
torch.save(state, path)
This approach is problematic because:
- The last model is not always the best model.
- The script could fail before the end of a training job.
Determined checkpointing is quite robust:
- Periodically during training, to keep a record of the training progress.
- During training, to enable recovery of the trial in case of resumption or errors.
- Upon completion of the trial.
- Prior to the searcher making a decision based on the validation of trials, ensuring consistency in case of a failure.
4) Hyperparameter search
Running a hyperparameter search is as simple as configuring them in your config file like so:
name: retinamnist_const
hyperparameters:
global_batch_size: 128
data_flag: retinamnist
lr: 0.001
weight_decay:
type: log
base: 10
minval: -4
maxval: -1
beta1:
type: double
minval: 0.1
maxval: 0.999
beta2:
type: double
minval: 0.1
maxval: 0.999
gamma: 0.1
min_validation_period:
epochs: 1
records_per_epoch: 1080
searcher:
name: adaptive_asha
metric: test_loss
smaller_is_better: true
max_length:
epochs: 15
max_trials: 16
mode: aggressive
resources:
slots_per_trial: 2
entrypoint: model_def:MyMEDMnistTrial
max_restarts: 0
With half the lines of code, and more than 4x the functionality, Determined makes it much easier for you to experiment with training models, especially if you are a researcher who doesn’t want to write infrastructure. Not to mention, the WebUI makes it easy for you to collaborate with your colleagues by viewing experiment submissions, cluster usage, etc:
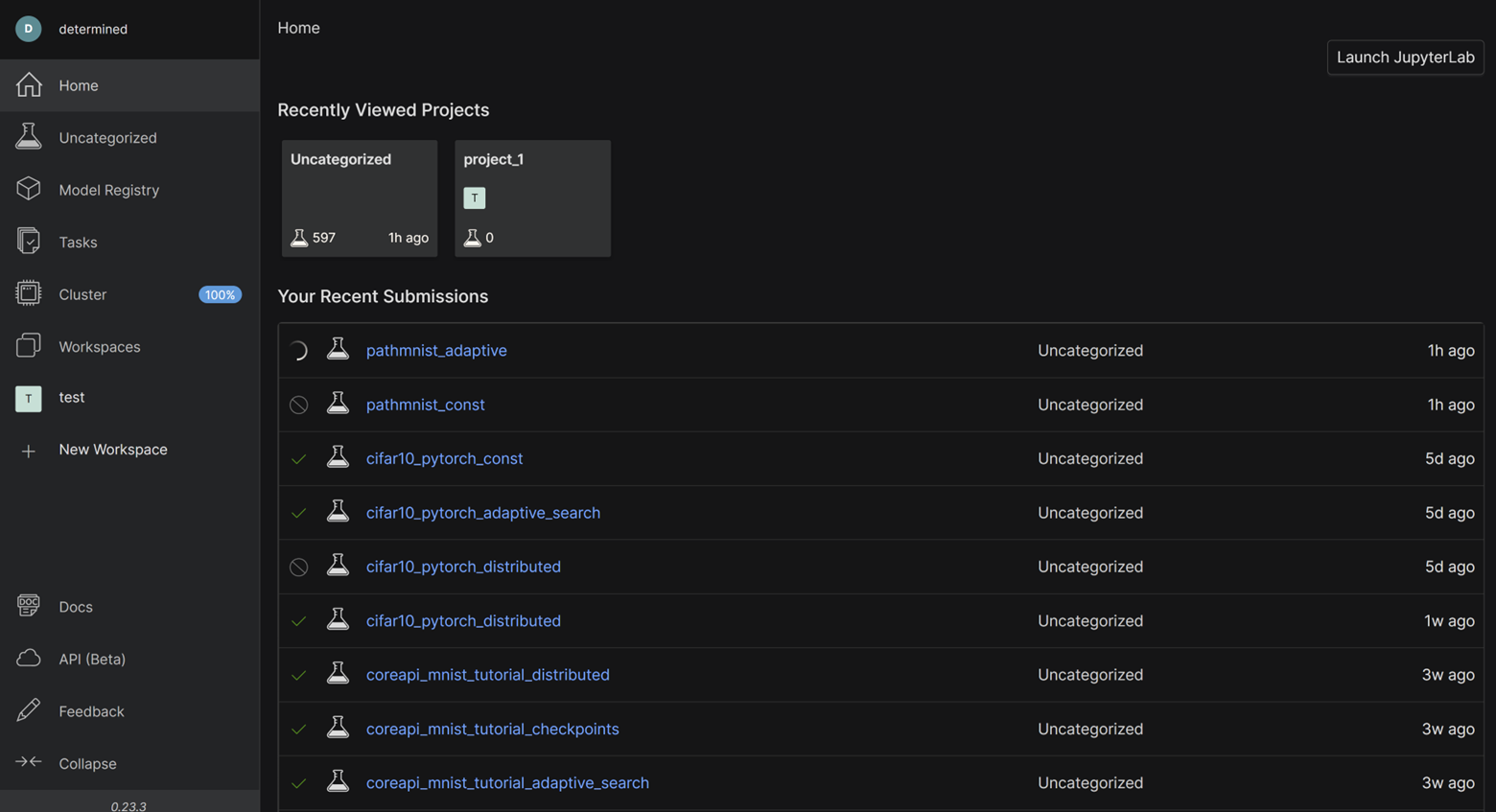
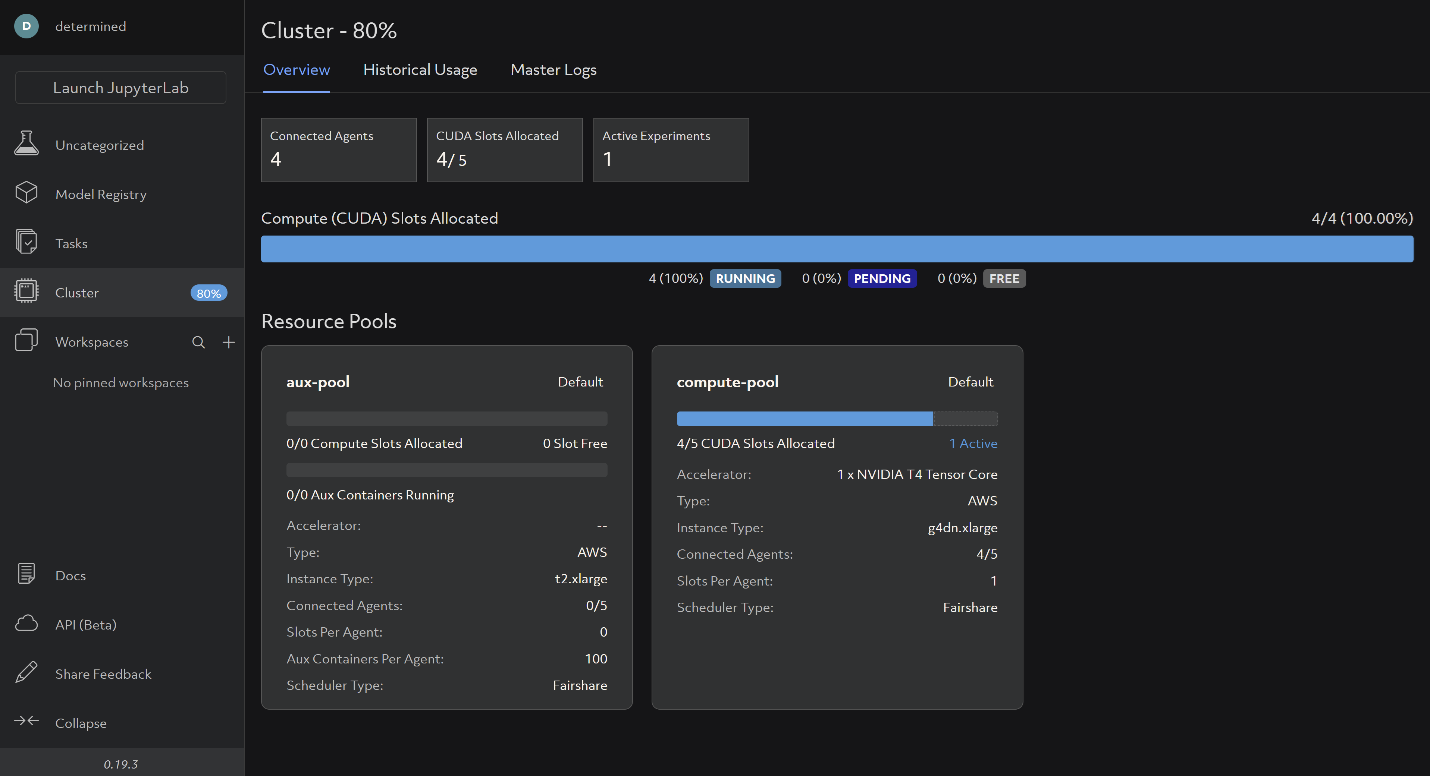
That’s it! We encourage you to jump into the code and try it out yourself using this blog as a guideline. Check out the GitHub repo and get in touch with us on the Determined Community Slack.
